 accesses since July 8, 1996
accesses since July 8, 1996 copyright notice
copyright notice
 link to published version: ACM Transactions on Information Systems (TOIS), January, 1996
link to published version: ACM Transactions on Information Systems (TOIS), January, 1996
 accesses since July 8, 1996
accesses since July 8, 1996Hal Berghel, University of Arkansas
David Roach, Acxiom Corporation
We describe an improvement on Ukkonen's Enhanced Dynamic Programming (EHD) approximate string matching algorithm for unit-penalty four edit comparisons. The new algorithm has an asymptotic complexity similar to that of Ukkonen's but is significantly faster due to a decrease in the number of array cell calculations. A 42% speedup was achieved in an application involving name comparisons. Even greater improvements are possible when comparing longer and more dissimilar strings. While the speed of the algorithm under consideration is comparable to other fast ASM algorithms, it has greater effectiveness in text processing applications because it supports all four basic Damerau-type editing operations.
Approximate string matching (ASM) refers to a class of techniques which associate strings of symbols with one another on the basis of some criteria of similarity. It is convenient to classify such similarity relations between strings by means of Faulk categories [1]: positional similarity (the degree to which matched symbols are in the same respective positions), ordinal similarity (whether they are in the same order) and material similarity (the degree to which they consist of the same symbols). Most modern ASM techniques use more than one type of similarity measure since single-measure techniques tend to be too narrow or too broad (for example, anagrams are materially identical, while 'fox' and 'ox' have nothing in common, positionally).
One historically significant multiple-relation similarity test is the Levenshtein Metric [2] which calculates the difference between strings in terms of an edit distance, or the minimum number of basic editing operations which can transform one string into the other. Typical basic editing operations are insertion, deletion, substitution and adjacent transposition. These particular operations are especially important in text processing applications [3] because they represent the most common forms of typing errors [4]. Such edit distance algorithms classify strings as similar when their edit distance is less than some threshold, k. Not surprisingly, this problem is commonly called the k differences problem in the literature. This problem is also known as the evolutionary distance problem in the context of molecular biology [5] and the string-to-string correction problem in text processing circles [6]. There are also several variations on this general theme, including the k mismatches problem which locates all occurrences of a string in a corpus which have no more than k mismatches. For excellent surveys of the underlying ASM algorithms, see Galil and Giancarlo [7], and Ukkonen [8].
With the dynamic programming approach, the edit or "Levenshtein" distance between two strings is represented as a path through a directed graph whose nodes d(i,j) are connected with horizontal, vertical, and diagonal edges representing the penalty or 'cost' of an edit operation. For the edit operations insertion, deletion, substitution and adjacent transposition with unit penalties, the edit distance between strings s1 = c1c2...ci and s2=c'1c'2...c'j may be defined recursively as follows:
d(0,0) = 0
d(i,j) = min[d(i,j-1)+1,
d(i-1,j)+1,
d(i-1,j-1)+v(ci,c'j),
d(i-2,j-2)+v(ci-1,c'j)+v(ci,c'j-1)+1]
where,
v(ci,c'j)=0 <-> ci = c'j, and v(ci,c'j)=1 <-> ci =/= c'j,
and the boundary conditions are d(i,0) = i and d(0,j) = j.
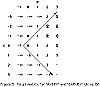
Figure 1 shows the d(i,j) array for determining the edit distance between the strings "AVERY" and "GARVEY".

While there are many interesting aspects of approximate string matching based upon edit distances, the current study focusses on the search for the shortest weighted path (i.e., the path with the least cumulative 'cost') with a minimum of computational effort. Since penalties are cumulative, the cost is inversely related to the degree of string similarity. The minimal path is identified by arrows in Figure 1. The value in cell d(5,6) is the edit distance, which in this case equals 3. (Unless otherwise noted, the basic edit operations of insertion, deletion, substitution, and adjacent transposition are assumed throughout).
The efficiency of the dynamic programming approach for determining edit distances was improved by Ukkonen [8][9]. Where the earlier algorithm of Wagner and Fisher [6] was implemented in time O(n²), Ukkonen's algorithm runs in O(s * n) time, for edit distance s and string length n. The improvement results from a reduction in the range of values in an (m+1) by (n+1) array (d(i,j)) that must be calculated in order to determine the edit distance (d(m,n)).
Ukkonen observed that the d(i,j) values form a non-decreasing sequence along any given diagonal, i.e.,
For every i,j, d(i,j) - 1 <= d(i-1,j-1) <= d(i,j).
On this account, the kth diagonal, for -m <= k <= n, consists of the sequence of d(i,j) cells for which j-i = k. Ukkonen's observation is that we need only determine those d(i,j) values, p, for which i is the highest numbered row in which p occurs on diagonal k (for a restricted range of k to be specified). The value i is a function of k and p such that
f(k,p) = the largest index i such that d(i,j) = p and d(i,j) is on diagonal k.
Consequently, the edit distance d(m,n) is equal to p such that f(n-m,p) = m.
Wu, et al, [10] improved on Ukkonen's basic approach by further restricting the range of d(i,j) values calculated. Let a and b be two strings of length m and n respectively, where n >= m, s is the edit distance between them, and p = ½s - ½(n-m). This algorithm has a worst case running time of O(n * p) and an expected running time of O(n + (p * s)). When a is a subsequence of b the running time is O(n). This time complexity is accomplished by evaluating only the narrow band of values in the d(i,j) array from -p to (n-m)+p. However, the only edit operations recognized by this algorithm are insertion and deletion, which makes it impractical for many text processing applications.
The algorithm which we present in the remaining sections has time complexity and experimental efficiency comparable to the algorithm of Wu, et al. However, it has a very different operational behavior and illustrates yet another way that the number of computed penalty array cells can be reduced. Additionally, it incorporates a wider range of edit operations including substitution and adjacent transposition. The algorithms of Wagner and Fischer; Ukkonen; and the authors are referred to as WF, UK, and BR, respectively, in the discussion to follow.
The kernel of algorithm UK which with minor modifications is shared by BR computes f(k,p) for strings a = a1,...,am and b = b1,...,bn. It is recursively defined as follows:
t := f(k,p-1) + 1;
t2 := t;
if atat+1 = bk+t+1bk+t then
t2 := t+1;
t := max(t, f(k-1,p-1), f(k+1,p-1) + 1,t2);
while at+1 = bt+1+k and t < min(m,n-k) do
t := t+1;
f(k,p) := t;
There are two terminating conditions on the recursion:
1) if p = |k| - 1 then
if k < 0 then
f(k,p) := |k| - 1;
else
f(k,p) := -1;
endelseif
endif
2) if p < |k|- 1 then
f(k,p) := -inf.
endif
The iterative version requires a array for holding f(k,p) values. We define a two-dimensional array (called FKP) having MAX_K rows whose indices correspond to d(i,j) array diagonal numbers and MAX_P columns whose indices range from -1 to the largest possible d(i,j) array cell value. The f(k,p) array cell values represent rows in the d(i,j) array. f(k,p) array row MAX_K/2 (referred to as row ZERO_K) always corresponds to the 0th diagonal of d(i,j). By 'anchoring' the 0th d(i,j) array diagonal to row ZERO_K of the f(k,p) array, a one-time initialization of the f(k,p) array is all that is required before successive string comparisons. The initialized cells are unaffected as strings of differing lengths are compared. Those cells are initialized in which the relationship between k and p is as defined by the recurrence terminating conditions. The following algorithm performs the necessary initialization.
for k := - ZERO_K to MAX_K - ZERO_K
for p := -1 to MAX_P-2
if p = |k| - 1 then
if k < 0 then
FKP[k+ZERO_K,p] = |k| - 1;
else
FKP[k+ZERO_K,p] = -1;
endelseif
else if p < |K| then
FKP[k+ZERO_K,p] = -inf.
endelseif
endfor
endfor
(The areas to the left of the arrows in later f(k,p) array figures are the initialized areas.) Notice that algorithms UK, WM, and BR operate on an f(k,p) array rather than the d(i,j) array used in the original dynamic programming algorithm (WF). However, since each f(k,p) array has a d(i,j) counterpart which is often more readily understandable we will often compare the output of the algorithms in terms of their d(i,j) arrays.
The recursive evaluation of f(k,p) values in UK's kernel is changed to simple array lookups in BR.
t := FKP[k+ZERO_K,p-1] + 1;
t2 := t;
if atat+1 = bk+t+1bk+t then
t2 := t+1;
t := max(t, FKP[k-1+ZERO_K,p-1],
FKP[k+1+ZERO_K,p-1] + 1,t2);
while at+1 = bt+1+k and t < min(m,n-k) do
t := t+1;
FKP[k+ZERO_K,p] := t;
As noted by Ukkonen [8][9], the range of f(k,p) values calculated can be further reduced by incorporating into the algorithm the observation that a necessary (but not sufficient) condition for d(i,j) being on a minimizing path from d(0,0) to d(i',j') is that
(1) d(i',j') >= d(i,j) + |j'-i'-(j-i)| for i <= i', j <= j'
If we let d(i',j') equal the edit distance d(m,n) then
(2) d(m,n) >= d(i,j) + |n-m-(j-i)|
The proof that (1) states a necessary condition for cell (i,j) being on a minimizing path is found in [9]. It is possible to construct an algorithm which never evaluates any d(i,j) that does not satisfy (1) even in a worst case comparison where the edit distance s = d(m,n) = max(m,n).
In order to more clearly relate (1) and (2) to the algorithm, we can restate them in terms of k, p, and s, beginning with (2). Clearly, d(m,n) = s and d(i,j) = p for f(j-i,p) = i. |n-m-(j-i)| represents the distance between the diagonal n-m on which d(m,n) lies and the diagonal j-i on which d(i,j) lies. We can let the (j-i)th diagonal be identified by k. Thus, the formula becomes
(3) s >= p + |n-m-k| (restatement of (2))
The Ukkonen formulation shows that the relation also holds between a given p and certain other values p' < s, i.e.,
(4) p' >= p + |k'-k| where k' is the diagonal associated with p'
(restatement of (1))
The formula restricts the range of p for a given k beyond that of UK. As a result, BR, which enforces the formula's constraints, calculates on the average fewer, and never more f(k,p) values than UK. Algorithm BR is as follows:
p := k;
repeat
inc := p;
for temp_p := 0 to p-1
if |(n-m) - inc| <= temp_p
f((n-m)-inc,temp_p);
endif
if |(n-m) + inc| <= temp_p
f((n-m)+inc,temp_p);
endif
inc := inc - 1;
endfor
f(n-m,p);
p := p + 1;
until FKP[(n-m)+ZERO_K,p-1] = m;
s := p-1;
procedure f(k, p)
begin
t := FKP[k+ZERO_K,p-1] + 1;
t2 := t;
if atat+1 = bk+t+1bk+t then
t2 := t+1;
t := max(t, FKP[k-1+ZERO_K,p-1],
FKP[k+1+ZERO_K,p-1] + 1,t2);
while at+1 = bt+1+k and t < min(m,n-k) do
t := t+1;
FKP[k+ZERO_K,p] := t;
end
As the outer repeat loop increases p (the array column index), the for loop increases the range of array rows whose cell values are calculated to ensure that for 0 <= i <= p, k is within i of f(n-m,p-i), with the restriction that p >= |n-m|. BR is correct with respect to UK since it ensures that f(k,p-1), f(k-1,p-1), and f(k+1,p-1) are computed before evaluating f(k,p) and terminates only when f(n-m,p)=m. This also satisfies Ukkonen's minimizing path relation (4) and thus (3).
Comparison of UK and BR
We can obtain a formula for the number of f(k,p) array cells whose values are calculated by segmenting the array into regions, ascertaining the number of filled cells for each region, and obtaining a sum (cf. the core (darkened) cell values in Figures 3 and 5, below).
First, the row of the f(k,p) array which corresponds to diagonal k = n-m in the Levenshtein array will contain (s-(n-m)+1) filled cells. There will also be n-m other rows in the f(k,p) array with the same number of filled cells. Thus, this portion of the array contains (s-(n-m)+1) * (n-m+1) filled cells.
The remaining upper and lower portions of the array will contain rows of cells definable by the progressions 1,3,5,... or 2,4,6,.... In the case of the odd progression, the number of rows in each of the upper and lower regions will be ((s-(n-m))/2. In the case of the even progression, the number of rows in the upper and lower regions will be ((s-(n-m)-1)/2. The number of cells in these regions is thus determinable by the arithmetic series formula R/2 * (2a + d(R-1)) where R in this case is the number of rows defined above, a is the starting value in the progression and d is the increment. For the odd progression, there are therefore R² cells above and below or 2R² total cells. For the even case, there are R²+R cells in each region, or 2(R²+R) in total.
Substituting for R our formulas for the number of rows, above, we have (s-(n-m))² / 2 cells in the odd case, or ((s-(n-m))² -1)/ 2 cells in the even. If we ignore division remainders in the even case, we may generalize the formula (s-(n-m))² / 2 for both cases.
The total number of cells filled is then ((s-(n-m))² / 2) + ((s-(n-m)+1) * (n-m+1)). This simplifies to (((s² - (n-m)²) / 2) + s + 1). When string a is a subsequence of string b, the algorithm is linear since only n-m+1 cells are filled.
The formula for Ukkonen's algorithm can be obtained in a related manner. Assuming that n=m, the number of cells filled is simply (s+1)² {-m if s = n, since the final column is not completely filled when s = n}. This is due to the symmetry of the fill region about row n-m. However, in the cases where n =/= m, the fill region becomes asymmetrical and we must subtract from the total of filled cells the number that are on rows too far from row n-m to be considered by the algorithm. The number of cells in this region of the array is ((n-m)² + (n-m)) / 2. Thus the number of cells calculated by UK is (s+1)² - (((n-m)² + (n-m)) / 2) {-m if s=n}. A comparison of these formulas suggests that BR would be considerably faster. This is evidenced by the difference in the dominant factors, s²/2 for BR versus (s+1)² for UK.
Figure 2 overlays the d(i,j) arrays that result from a comparison of "ABCDE" and "FGHIJ" using algorithms UK and BR. The outlined values are those calculated by UK but not BR.

As shown in figure 3, BR calculates fewer cells since it does not calculate cell values as far off diagonal n-m as UK for higher values of P.

UK was tested on a database of 5,000 randomly selected last name pairs. (This work was motivated by a need to identify duplicate name/address records with corruptions.) Table 1 lists some sample names and edit distances.
Table 1. Sample last name pairs and edit distances. Name 1 Name 2 Edit Distance ADCROFT ADDESSI 5 BAIRD BAISDEN 3 BOGGAN BOGGS 2 CLAYTON CLEARY 5 DYBAS DYCKMAN 4 EMINETH EMMERT 4 GALANTE GALICKI 4 HARDIN HARDING 1 KEHOE KEHR 2 LOWRY LUBARSKY 5 MAGALLAN MAGANA 3 MAYO MAYS 1 MOENY MOFFETT 4 PARE PARENT 2 RAMEY RAMFREY 2
Iterative versions of WF, UK and BR were run on the same data. Of course, all three algorithms calculated the same edit distances. However, there are significant differenc es in the resulting run-times. Table 2 shows the relative run-times and percentage improvements of BR over UK.
Table 2. Summary of timings in system clock ticks. Algorithm Time Speedup (wrt WF) Speedup (wrt UK) WF 90 - - UK 33 63% - BR 19 79% 42%
UK is 63% faster than WF. BR is 42% faster than UK making it 79% faster than WF. Of course, these improvements are accomplished with no loss of accuracy.
Figure 4 contains the combined UK and BR d(i,j) arrays resulting from a comparison of the strings "AVERY" and "GARVEY". It illustrates how fewer d(i,j) values are calculated by BR for the sample strings.

Figure 5 shows the corresponding f(k,p) arrays. As the region to the right of the arrows illustrates, BR converges toward d(m,n) (p = 3) by focusing on the diagonal n-m. In contrast, UK diverges by considering an increasing range of k for higher values of p.
We point out that the improvement in efficiency is directly proportional to the decrease in the number of f(k,p) values calculated, because procedure f(k,p) is called each time an f(k,p) value is to be obtained.
The efficiency of our algorithm results from (1) an iterative implementation that re quires a one-time initialization of an f(k,p) array and (2) a more restricted range of calculated d(i,j) values based on a minimizing path formula. Performance comparisons of iterative versions of the basic dynamic programming algorithm, Ukkonen's algo rithm and ours reveal that ours is 42% faster than Ukkonen's. Further, it is comparable in efficiency to the two-edit algorithm of Wu et al while more effective in VLDB applications involving name comparisons which require the support of a broader range of basic edit operations.
This work was supported in part by matching grants from the Acxiom Corporation and the Arkansas Science and Technology Authority. We wish to thank David Andrews, Gordon Beavers, Daniel Berleant, Ed Fox, Bernard Moret, Esko Ukkonen and several anonymous referees for many useful comments on earlier drafts of this paper.
Two new ASM algorithms have been reported in the literature since the submission of this manuscript. While they do not alter the results reported above, they do change the perspective in which our results should be placed.
First, an alternative formulation of our algorithm, one which uses two nested for loops instead of a conditional test within one for loop, was suggested by one of the anonymous reviewers. While slightly slower in our application because the overhead of the for loop exceeds the overhead of the conditional statements, it is nonetheless an interesting alternative. Its behavior is perspicuous and it cleverly anticipates the distance that p-i extends down the diagonals n-m±i. It should be noted that while this algorithm calculates the same f(k,p) values as our algorithm, it does so in a different order.
Here is the algorithm as provided by the reviewer. Assume that n>=m,
p := n-m-1;
repeat
p:= p+1;
for i := (p-(n-m)) div 2 downto 1 do
f(n-m+i,p-i);
for i:= (n-m+p) div 2 downto 1 do
f(n-m-i,p-i);
f(n-m,p)
until FKP(n-m+ZERO_K,p)=m
Second, Wu and Manber [11] reported a new 3-edit, k-mismatch algorithm which has performance similar to ours and Ukkonen's. While it is only a 3-edit algorithm, it appears to be easily extensible to a wide variety of input formats and may be extended in a limited way to accommodate non uniform mismatch penalties. It is currently embedded in an extension of the Unix utility, grep.
In addition to these two new algorithms, we have developed an interactive, algorithmic animator which demonstrates the behavior of Ukkonen-style ASM algorithms. The animator simultaneously displays the f(k,p) and d(i,j) (or "Levenshtein") arrays as they are filled by the algorithms reported in this paper. It also features a "step-through" toggle which activates the display of the current variable assignments, the temporary stack contents and string position pointer locations as the algorithm proceeds. The program, called "the String Thing", is available via anonymous ftp at
cavern.uark.edu /people/hlb/prototypes/approx_string_matching
or through WWW via the first author's home page at
http://berghel.net
by selecting the "FTP site" link. The String Thing, which runs under DOS 4.0 or higher, Windows and OS/2, comes in three executable versions and compares various combinations of ASM algorithms mentioned in this article.
[1] Faulk, R., An Inductive Approach to Language Translation. Communications of the ACM, 7 (1964) 647-653.
[2] Levenshtein, V. Binary Codes Capable of Correcting Deletions, Insertions and Reversals. Sov. Phys, Dokl., 10 (1966), 707-710.
[3] Berghel, H. A Logical Framework for the Correction of Spelling Errors in Electronic Documents. Information Processing and Management, 23 (1987) 477-494.
[4] Damerau, F. J. A Technique for Computer Detection and Correction of Spelling Errors. Communications of the ACM, 7 (1964) 171-176.
[5] Sellers, P. On the Theory and Computation of Evolutionary Distances. SIAM J. of Applied Math. 26 (1974), 787-793.
[6] Wagner, R. & Fischer, M. The String-to-String Correction Problem. Journal of the ACM, 21 (1974),168-178.
[7] Galil, Z. and R. Giancarlo. Data Structures and Algorithms for Approximate String Matching. Journal of Complexity 20 (1988), 33-72.
[8] Ukkonen, E. Algorithms for Approximate String Matching. Information and Control 64 (1985), 100-118.
[9] Ukkonen, E. On Approximate String Matching. Proceedings of the International Conference on Foundations of Computation Theory, Borgholm, Sweden. (August, 1983). 21-27.
[10] Wu, S., Manber, U., Myers, G., & Miller, W. An O(NP) Sequence Comparison Algorithm. Information Processing Letters 35 (1990), 317-323.
[11] Wu, S. and U. Manber. Fast Text Searching Allowing Errors. Communications of the ACM 35 (1992), 83-91.